SPM Lesion Normalization with Source Weighting Image¶
Goal: Warp a native space lesion mask (and optionally a native space anatomical image) into standard (MNI) space. In this procedure we use an inverse lesion mask to define areas that should influence the warp. The old normalize procedure did not depend on an initial segmentation and thus had distinct disadvantages over the newer registration that matches tissue type boundaries.
Data¶
- Images must not be zipped (i.e., T1w.nii is okay, but T1w.nii.gz is not).
- An anatomical (anatomical) image, usually T1 weighted, but could be T2 or flair weighted.
- A lesion mask with values of 1 in the lesion and 0 elsewhere.
Steps¶
Create Inverse Lesion Mask¶
If you have a mask with 1 anywhere there is lesion and 0 elsewhere, you need the inverse of that. To create this inverse mask in SPM12, you can use the image calculator (N.B. Although the resulting mask displayed in FSL shows the “1” to be slightly more or less than 1, SPM nevertheless seems to treat the result as a true binary mask). You have two choices for accessing the image calculator:
- Choose ImCalc from the main SPM menu
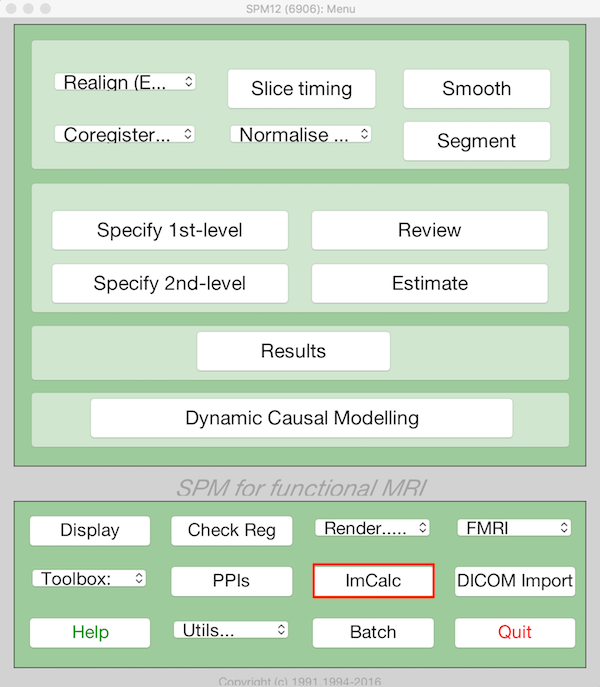
- Select Batch and from the menubar: SPM ➜ Util ➜ Image Calculator
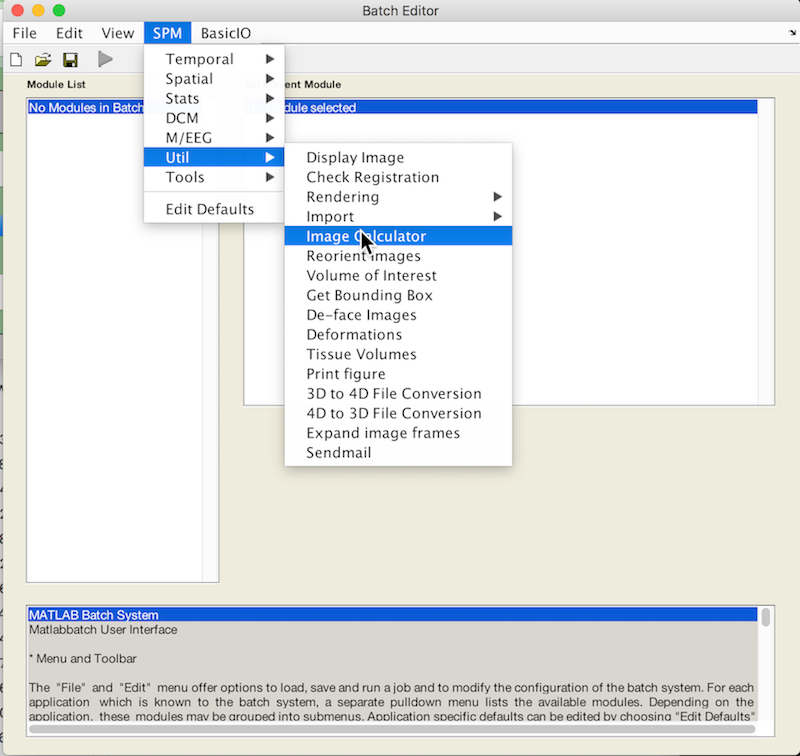
- An X is displayed for the two parameters that must be specified: Input Images and Expression. You may also change the Output filename and directory.
- The Input Image will be the native space lesion mask
- The expression will be (i1 .* -1)+1
- Set Interpolation to Nearest Neighbor to prevent values between 0 and 1 from being created at the edges of the lesion.
- Set Data Type to UINT8 -unsigned char
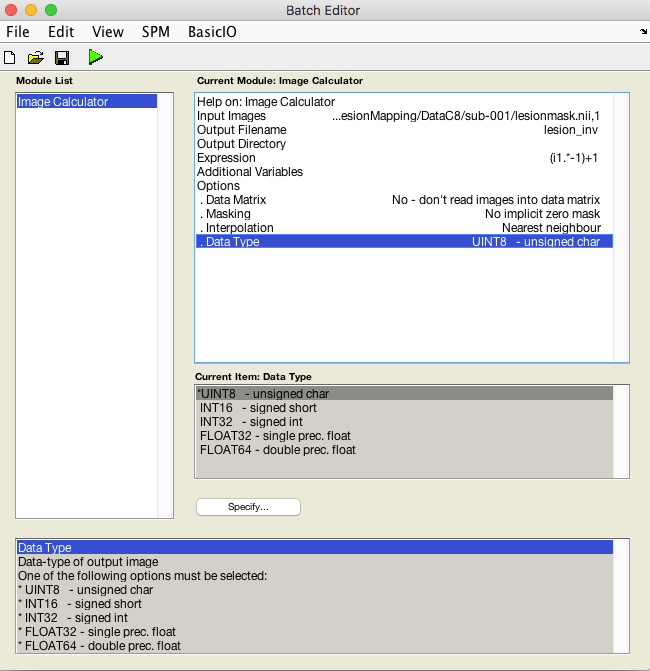
- In the image calculator, i1 always refers to the first input image you selected (i2= the second image etc.). In this expression we multiply all values in i1 by -1, so 0 remains 0, but 1 becomes -1. This inverts the values.Then we add 1 to all voxels the resulting image so that the range is 0 to 1 and the values have been inverted.
- This is the first module of your batch.
Old Normalize: Estimate and Write Lesion Mask¶
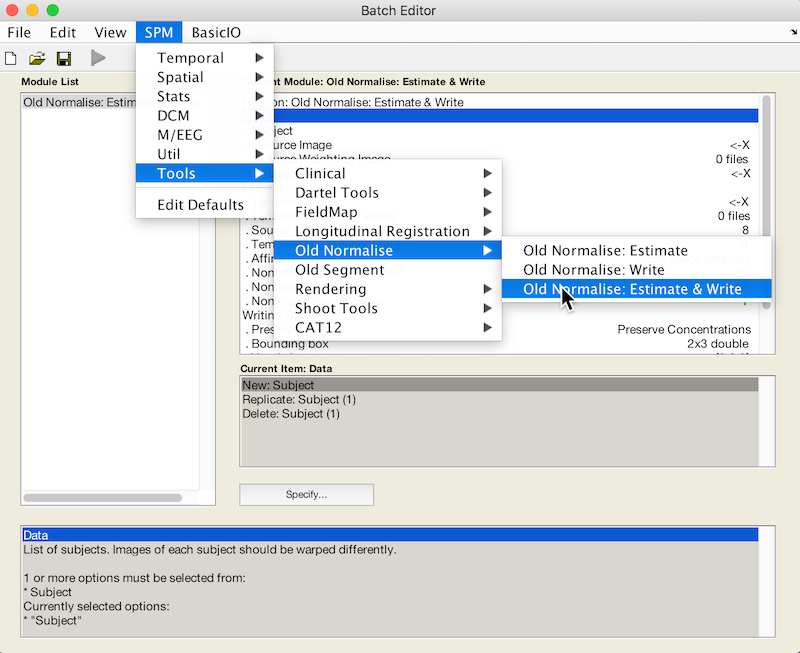
- Select Batch and from the menubar: SPM ➜ Tools ➜ Old Normalise ➜ Old Normalise: Estimate and Write
- Click Data, and select New Subject from the grey Current Item: Data box below. X values are displayed, indicating parameters that must be specified.
- Specify the native space anatomical image that you used to draw the lesion as the Source Image. * We also want a Source Weighting Image. Click Source Weighting Image and select the Dependency button. Choose the Image Calculator: ImCalc Computed Image for your inverse lesion file.
- For Images to Write select your original lesion file.
- Under Estimation Options Select a Template Image from the OldNorm subdirectory, choose a template with the same contrast as your source image (i.e., if our source image is a T1, choose T1.nii.)
- Under Writing Options find Interpolation and select Nearest neighbour
- This is the second module of your batch.
Old Normalize: Write Anatomy file¶
- From the batch editor, choose: SPM ➜ Tools ➜ Old Normalise ➜ Old Normalise: Write
- For the Parameter file select Dependency and the only Norm Params File available.
- Select the anatomical file for Images to Write
- Leave the default trilinear interpolation for this one.
- All other values should be left as their defaults.
Run Batch and View Results¶
- Run the batch module by clicking the green go button (upper left on batch editor).
- The ImCalc module will generate the inverse lesion mask.
- Normalization produces warped files prefixed with w.
- In addition, a _sn.mat file is produced that can be used to write additional warped images.