bids-validator¶
Introduction¶
BIDS datasets should conform to standards described in the evolving Brain Imaging Data Structure (BIDS) Specification. When you run a BIDS app, the data structure is checked to ensure it conforms (though BIDS apps normally allow you to turn the checking off).
bids-validator online¶
You can check your data structure using the online bids-validator (which works in Google Chrome and Mozilla Firefox). At the top of the interface where it says Choose File, select the parent directory containing subject subdirectories. There must be at least one subject directory for the validator to run correctly. Don’t worry!! You are not uploading your files, the validator is looking at the directory and image names and the json file names and contents.
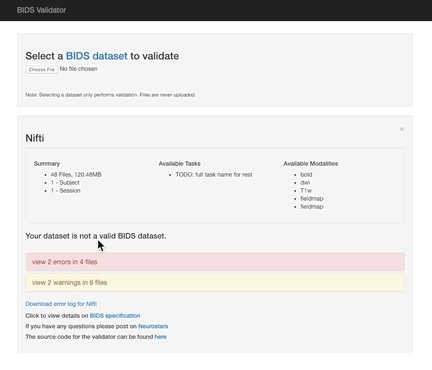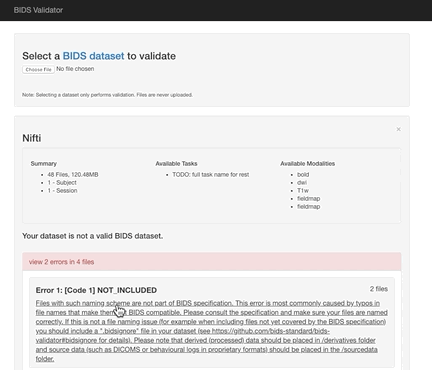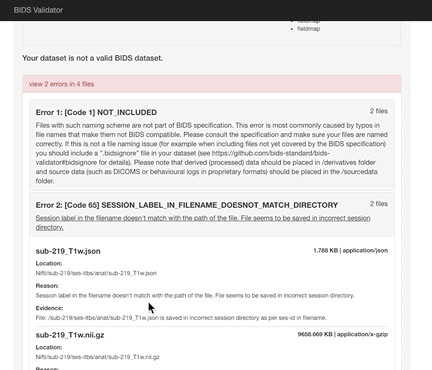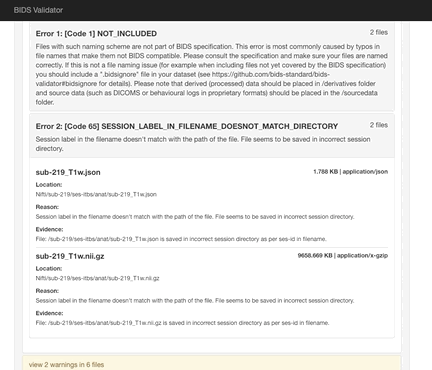
Here the error is that any image and json file in a session directory must include the session label in its name. Error messages from the validator can be very helpful, but are sometimes nonsensical (e.g., claiming that you can’t use hyphens or underscores or that a field does not exist in a json file, even though you know it does exist). If nonsensical, you should should not take the reported infractions at face value, they likely reflect a different problem.
- Review the naming carefully and ensure that you don’t have any directories containing non-bids data in the path.
- If you use sessions, then every image under a session directory must be labelled with the session label.
- Also click Download error log for Nifti (see bottom of bids-validator page) so that you can submit the log to the BIDS group for help.
Phasediff images¶
Some BIDS apps (e.g., MRtrix3_connectome) expect the phasediff .json sidecar to include EchoTime1 and EchoTime2 fields. dcm2niix does not currently provide those two fields because of the way Siemens has stored the dicom information. The phasediff image gets exported with a single EchoTime field (which actually corresponds to EchoTime2). You can find EchoTime1 in the Magnitude1 image (again, just labelled EchoTime). However, at this point you have to manually alter the phasediff json file to include the correct fields and values for apps that require it (e.g., MRtrix3_connectome). A single .json file that contains the correct information can also be added at the level of the parent directory.
BIDS apps with narrow assumptions¶
Sometimes BIDS apps assume naming conventions which are not required for bids validation. For example, the app may expect particular fields in the json file or a particular naming convention for the files rather than handling the variety of possibilities acceptable to the validator.
In addition, certain kinds of processing (like running FSL topup) rely pretty exclusively on MRI data from a particular manufacturer (Siemens). So, despite the goal of using any BIDS app on any correctly organized dataset, there may still be problems for a particular app with special assumptions.
Creating a .bidsignore¶
If you are including files or directories that are not handled by the official bids specification, then the bids-validator will produce errors. To address this issue, you can create a small hidden file at the same level as your subject directories. .bidsignore uses the syntax of .gitignore
For example, this tells the bids-validator to ignore any files in any directories that contain asl or tse in the names:
**/*asl*
**/*tse*
This is useful since (as of this writing) the asl and tse filetypes are not yet handled by BIDS. The .bidsignore file is directly under the directory Nifti in the example below.
Example¶
The directory structure below is fully bids compliant. It does not have to be called Nifti, but that is the default used by HeuDiConv:
Nifti
|-- .bidsignore
|-- .heudiconv
| `-- 219
| |-- info
| | |-- 219.auto.txt
| | |-- 219.edit.txt
| | |-- convertall.py
| | |-- convertall_old.py
| | |-- dicominfo_wrong_name.tsv
| | `-- filegroup.json
| `-- ses-itbs
| `-- info
| |-- 219_ses-itbs.auto.txt
| |-- 219_ses-itbs.edit.txt
| |-- convertall.py
| |-- dicominfo_ses-itbs.tsv
| `-- filegroup_ses-itbs.json
|-- CHANGES
|-- README
|-- code
| |-- __pycache__
| | `-- convertall.cpython-36.pyc
| `-- convertall.py
|-- dataset_description.json
|-- participants.tsv
|-- sub-219
| `-- ses-itbs
| |-- anat
| | |-- sub-219_ses-itbs_T1w.json
| | |-- sub-219_ses-itbs_acq-tse_T2w1.json
| | |-- sub-219_ses-itbs_acq-tse_T2w1.nii.gz
| | |-- sub-219_ses-itbs_acq-tse_T2w2.json
| | |-- sub-219_ses-itbs_acq-tse_T2w2.nii.gz
| | `-- sub-219_ses_itbs_T1w.nii.gz
| |-- dwi
| | |-- sub-219_ses-itbs_acq-AP_dwi.bval
| | |-- sub-219_ses-itbs_acq-AP_dwi.bvec
| | |-- sub-219_ses-itbs_acq-AP_dwi.json
| | `-- sub-219_ses-itbs_acq-AP_dwi.nii.gz
| |-- fmap
| | |-- sub-219_ses-itbs_dir-PA_epi.json
| | |-- sub-219_ses-itbs_dir-PA_epi.nii.gz
| | |-- sub-219_ses-itbs_magnitude1.json
| | |-- sub-219_ses-itbs_magnitude1.nii.gz
| | |-- sub-219_ses-itbs_magnitude2.json
| | |-- sub-219_ses-itbs_magnitude2.nii.gz
| | |-- sub-219_ses-itbs_phasediff.json
| | `-- sub-219_ses-itbs_phasediff.nii.gz
| |-- func
| | |-- sub-219_ses-itbs_acq-asl_run-01.json
| | |-- sub-219_ses-itbs_acq-asl_run-01.nii.gz
| | |-- sub-219_ses-itbs_acq-asl_run-02.json
| | |-- sub-219_ses-itbs_acq-asl_run-02.nii.gz
| | |-- sub-219_ses-itbs_task-rest_run-01_bold.json
| | |-- sub-219_ses-itbs_task-rest_run-01_bold.nii.gz
| | |-- sub-219_ses-itbs_task-rest_run-01_events.tsv
| | |-- sub-219_ses-itbs_task-rest_run-02_bold.json
| | |-- sub-219_ses-itbs_task-rest_run-02_bold.nii.gz
| | `-- sub-219_ses-itbs_task-rest_run-02_events.tsv
| `-- sub-219_ses-itbs_scans.tsv
`-- task-rest_bold.json