Image Processing Tips and Tricks¶
Most of the tools described below are based on FSL. FSL is not the only tool that can be used for this sort of processing, it is just the tool I know best. Where appropriate, bash scripts are available for download from my Bitbucket tools site. You can just download the whole repository if you want, but it’ll include scripts that may be of no interest. Otherwise, the scripts you want are described in each section (left click a link to view the script, right-click to download). Also, see the Clinical Scans page for issues related to processing data from clinical scans (which may be CTs and or have very low resolution in two of the three planes).
Anatomical Image Preparation¶
Strutural images, usually T1-weighted, but sometimes T2-weighted, FLAIR, scalar DWIs (B0, FA, MD etc.) or even CTs may be available as underlays to draw lesion masks on. CTs are a really special case and are discussed separately on the Clinical Scans page. T1w images can be processed with a single script described in Prepare T1w Image that reorients, bias-corrects, crops and even generates a brain mask. Several of these processing steps can also be handled separately as described in other sections below.
Prepare T1w Image¶
For a T1w image, you can use prep_T1w.sh to run fslreorient2std, robustfov, bias correction, and to create a brain mask for a T1w image. Note that prep_T1w.sh depends on FSL and on two additional scripts being present: fsl_anat_alt.sh and optiBET.sh:
prep_T1w.sh sub-001_T1w
Optionally, if you have a lesion mask in native space, you can ensure that it also gets cropped and reoriented by adding it to the prep_T1w.sh command:
prep_T1w.sh sub-001_T1w lesion
Check the brain mask! Large lesions, especially chronic lesions near the surface of the cortex, frequently result in a brain mask that does not have enough coverage. If all of your data consists of high quality research scans, then consider using SPM12 normalization before you delineate the lesion. That way, you can use the standard brain mask instead of generating a brain mask.
Running prep_T1w.sh creates a subdirectory named after your anatomical file, e.g., sub-001_T1w.anat. It also backs up your original files, e.g., sub-001_T1w_orig.nii.gz and optionally your lesion file if you processed it, e.g., lesion_mask_orig.nii.gz. The suffix _orig has been added to these original files. Your new cropped images will have the names that you originally passed in (e.g., sub-001_T1w.nii.gz and lesion_mask.nii.gz).
fslreorient2std¶
It never hurts to reorient the image to standard orientaion. Some viewer programs will show you the image in standard orientaion whether or not it is stored that way. Other image viewers will show you the image as it is stored. For the sake of consistency, ensure the image is stored in the same orientation as the MNI standard. You can try this test data to see fslreorient2std in action: sub-001_T1w.nii.gz (this is my head, so no worries about HIPPA, it is free to use):
fslreorient2std sub-001_T1w sub-001_T1w_reoriented
The command is harmless if the data is already correctly oriented. If you apply the above command to the ITK-SNAP tutorial data T1w_defaced.nii.gz, nothing changes.
Crop Neck with robustfov¶
Crop the anatomical images, thus removing the lower head and neck. This command will run fslreoreint2std if it has not already been done.:
robustfov -i sub-001_T1w_reoriented -r sub-001_T1w_cropped
Correct Extreme Anisotropy¶
Clinical images often have extremely anisotropic voxels. This is not well handled by tools like MRIcron. In addition, ITK-SNAP needs isotropic (or near isotropic) data to do good semi-automated segmentation. You can easily make voxels isotropic with flirt: A simple FSL-based script to facilitate creating isotropic images (anatomical images or masks) is available here.: iso.sh. To try the script out, download T1w_aniso.nii.gz:
iso.sh T1w_aniso 1
The above command takes T1w_aniso.nii.gz as input and reslices it to 1mm isotropic voxels. The output is called T1w_aniso_1mm.nii.gz. The output image can now be used in ITK-SNAP or MRIcron. Note that the image is just as blurry, but it has more slices in the z direction now. That means any mask you draw will be smoother.
Reslice Other Anatomicals into One Space¶
You can use the script reslice.sh to facilitate running FSL flirt:
reslice.sh anat_FLAIR anat_T1w
- This example reslices an image,
anat_FLAIR.nii.gzto be in the same space asanat_T1w.nii.gz. - The script can be used, for example, to ensure that images of several different modalities are in the same space so they can be used with 3D segmentation clustering in ITK-SNAP.
- Clinical CT Another useful application of reslice.sh is to apply gantry tilt correction (Use the gantry tilt corrected image as the reference image).
- Fusion Images In the section on fusion images, I walk you through using reslice.sh to create a very simple fusion image.
Warning
reslice.sh uses trilinear interpolation, which is fast and fine for anatomical images. However, it is NOT appropriate for reslicing masks.
Masks¶
A mask (such as a lesion mask) should be binary, that is, it should contain ONLY 0’s and 1’s. This is because statistical programs for evaluating lesion masks, along with various image math procedures often assume the values are 0’s and 1’s. So, we want to make sure the masks remain binary.
Some tools, like MRIcron, offer a smoothing option for masks. The problem is that smoothing may be accomplished by softening the edges which means that those values are interpolated (i.e., 1’s are replaced with values like 0.5, 0.3 etc). In addition, if you register your mask to a different image space you can introduce interpolated values if you fail to use Nearest Neighbour interpolation. You should go through the Hidden Problems Section below, before registering a mask. In this section, I offer several tools for improving masks without compromising their binary nature.
Ensure Masks are Binary
If you reslice a mask into a different space, you should do so with Nearest Neighbour interpolation. Otherwise, values at the edges of the mask may be interpolated between 1 and 0. Smoothing a mask may result in the same problem. To ensure masks are binary, use fslmaths:
fslmaths lesion -bin lesion -odt char
Check that a mask contains correct values (-M Mean of non-zero voxels should be 1. -S Standard deviation of non-zero voxels should be 0:
fslstats lesion -M -S
1.000000 0.000000
Erode¶
Remove islands and spikes by eroding the edges of the lesion mask. erode.sh is a bit less complex to call than the fslmaths command. In the command below, we do one iteration of the erosion and do it in 3D (default is 2D):
erode.sh lesion 1 3D
Warning
Be Careful with erosion! If you run too many iterations on a small mask, the mask will disappear! You might want to make a backup copy of your mask first.
Dilate¶
Fill tiny holes in the mask by dilating the edges of the lesion mask. dilate.sh is a bit less complex to call than the fslmaths command. In the command below, we do one iteration of the dilation and do it in 3D (default is 2D):
dilate.sh lesion 1 3D
Fill Holes¶
fslmaths has its own hole-filling algorithm. Simply add the appropriate flag to fslmaths. | -fillh : fill holes in a binary mask (holes are internal - i.e. do not touch the edge of the FOV) | -fillh26 : fill holes using 26 connectivity | In the command below we input the lesion_mask, fill the holes and output it with the same name. Finally, we ensure it is the correct datatype:
fslmaths lesion -fillh lesion -odt char
Cluster Count¶
It can be difficult to determine whether a lesion mask is one big volume or whether little islands have broken off. cluster_count.sh tells you how many separate clusters are in the mask, how big they are and the center of each one. In general, I can’t imagine why you’d want to keep clusters of less than about 10 voxels. In the command below we ask how many clusters are in lesion_mask.nii.gz and we set connectivity to 6.
Connectivity can be set at 6, 18 or 26, but always defaults to 26.
- If the connectivity is set to 6, then voxels only belong to a single cluster if their faces are in contact.
- If the connectivity is set to 18, then voxels belong to a single cluster if their faces OR edges are in contact.
- And, if the connectivity is set to 26 (the default if you don’t specify a number), then voxels belong to a single cluster if their faces, edges or corners are in contact.
So, setting the connectivity to 6 ensures the greatest possible number of clusters will be counted:
cluster_count.sh lesion 6
Cluster Clean¶
If there are several clusters, especially if there are several tiny clusters, you may wish to remove the smallest ones. cluster_clean.sh can remove islands without the dangers of erosion. Often you’ll find you have one gigantic cluster (many thousands of voxels) and several clusters of 1-10 voxels. You can safely remove the tiny ones with the cluster_clean.sh. A command like the following which removes any clusters smaller than 10 voxels from the mask image:
cluster_clean.sh lesion 10
As with cluster_count.sh, a third argument can be added to specify connectivity of 6 or 18. The default connectivity is 26:
cluster_clean.sh lesion 10 18
Smooth¶
Yes, this is what I warned you against, but with some care it seems to work in FSL and produce binary results. You may have to play with the -kernel gauss 0.1 setting a bit. This seems to do very gentle smoothing, but still dilates the mask (and larger kernel values are more extreme). One iteration of erosion afterwards seems to do the trick. I ran this smoothing three times, each time running on the output of the previous iteration, and then eroding the results. The effects were improved:
fslmaths lesion -kernel gauss 0.1 -fmean -bin lesion_smooth
erode.sh lesion_smooth 1 3D
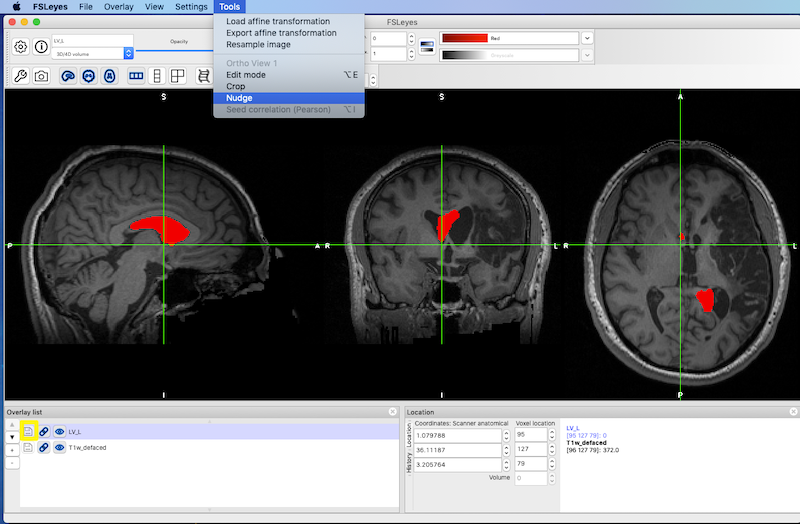
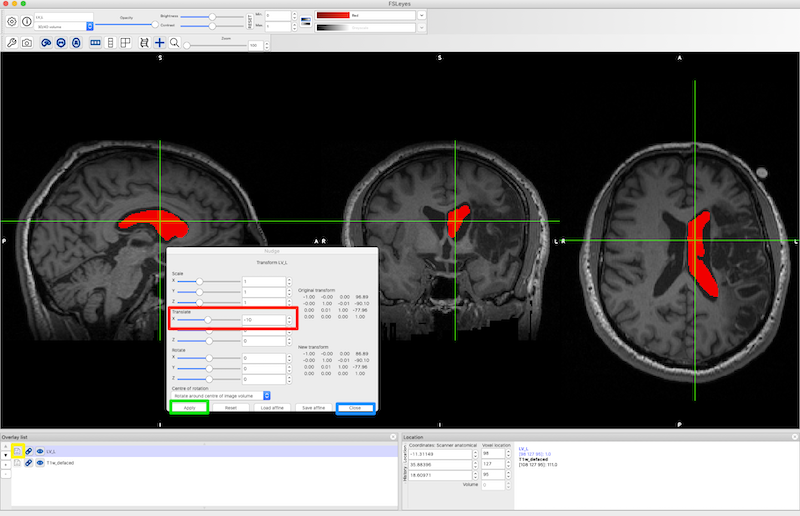
Left-Right Flip and Nudge¶
It can be useful to flip the normal ventricles in one hemisphere onto the abnormal ventricles in the other hemisphere to characterize ventricular enlargement. This is a two step process:
See Left-Right Flip and Nudging
Other¶
Edit Image Header (Mango)¶
If you encounter images with incorrect dimensions, you can revise the header using Mango Header Editing. This is handy if you have an image with the wrong slice thickness, for example.
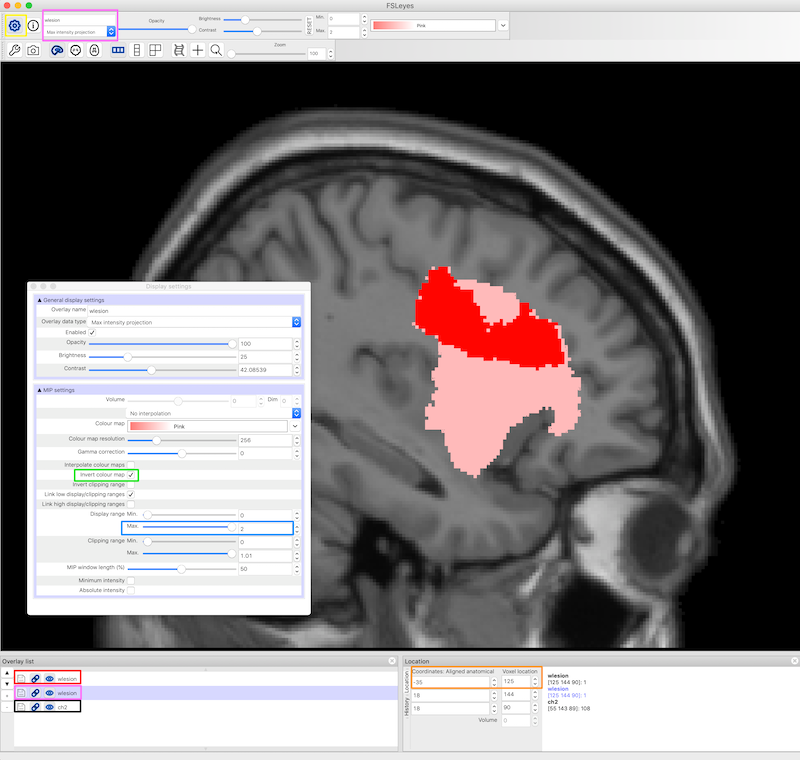
Visualization of Lesions using Maximum Intensity Projection¶
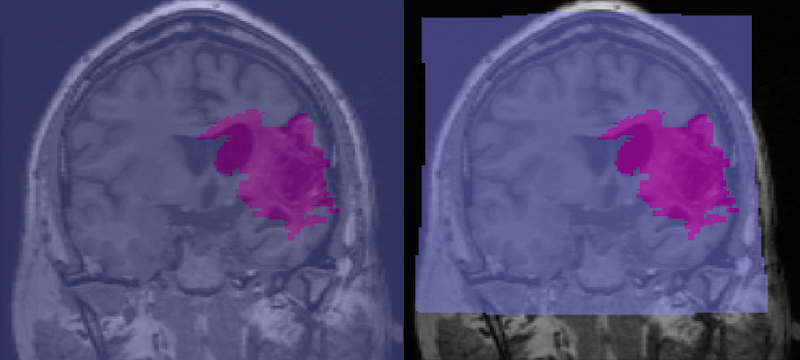
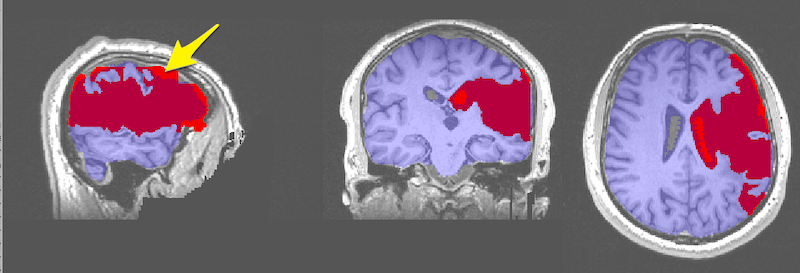
- In FSLeyes, load and display the anatomical image and two copies of the lesion file.
- By default the lesion file is displayed as a 3D/4D volume which displays the lesion mask for each slice. In the figure below, the 3D lesion is displayed in red (and has a red box around it in the Overlay list on the bottom left of the figure).
- Set the second lesion file to display as a Max Intensity Projection (MIP). It needs to be underneath the lesion file above, as it will probably be bigger. You can find the Max Intensity Projection in the upper left corner and the corresponding copy of the lesion file in the Overlay list (pink boxes, figure below). The MIP is also displayed in pink.
- Settings for the lesion maps are available from the gear on the upper left of FSLeyes (yellow box, figure below) and include an option to invert the colormap and otherwise refine the display. To get this particular shade of pink, we chose the pink color map, then Invert colour map (green box, figure below) and we set the Display range Max to 2. It also seems to work to use the default pink color map and set the max to about 4.
- We chose the ch2.nii.gz standard image, and set the x coordinates to -35 (orange box, figure below).
Convert Images from SPM MNI to FSL MNI¶
Not all MNI spaces are created equal. In 1 mm resolution, the SPM MNI image is 181x217x181 but the FSL MNI image is 182x218x182. This is not a lot, but is just enough to break some tools (like Tractotron) that expect one format and not the other. Here I provide a script to convert from the SPM12 MNI format to the FSL format. This script ensures that the shape, size and datatype of the image remain exactly the same, and, in fact, the coordinates of any structures remain exactly the same. I have Mark Jenkinson from the FSL group to thank for getting all the right flirt flags. You can download spm2fsl.sh
spm2fsl.sh lesion_mask_spm_1mm