ITK-SNAP¶
Introduction¶
ITK-SNAP is an image segmentation tool. ITK-SNAP is similar to MRIcron in that both are excellent tools for creating lesion masks on MRI images. ITK-SNAP may have some advantages: smarter fill tools (adaptive paintbrush and automatic segmentation) in addition to 3D editing tools. Because MRIcron assumes isotropic voxels (or near isotropic), it is not well suited to drawing on clinical data which often has high resolution in-plane but very thick slices. However, it is worth noting the ITK-SNAP needs isotropic (or near isotropic) data to do good semi-automated segmentation. Here is a simple FSL-based script to create isotropic images: iso.sh.
See the ITK-SNAP Segmentation of Stroke-related Lesions page for a link to some example data and instructions for using automatic segmentation for filling the lesion.
Load and View the Image / Images¶
- Locate the ITK-SNAP icon and double-click it.
- Select File ➜ Open Main Image from the menu.
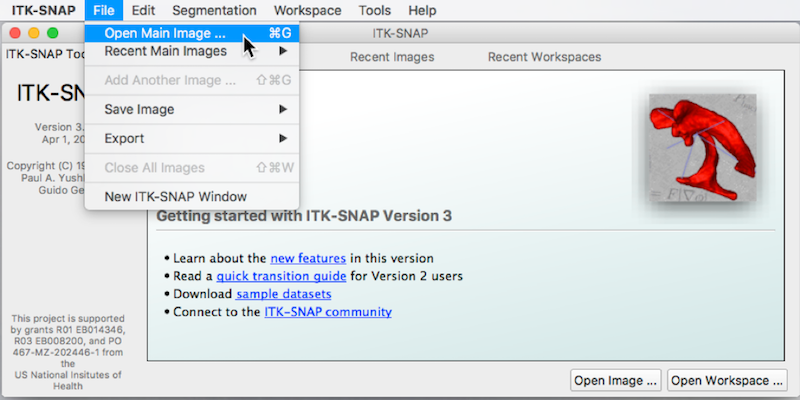
- An Open Image window appears (See figure below).
- Press Browse to search for the MRI image ➜ Select the image T1w_defaced.nii.gz and press Open.
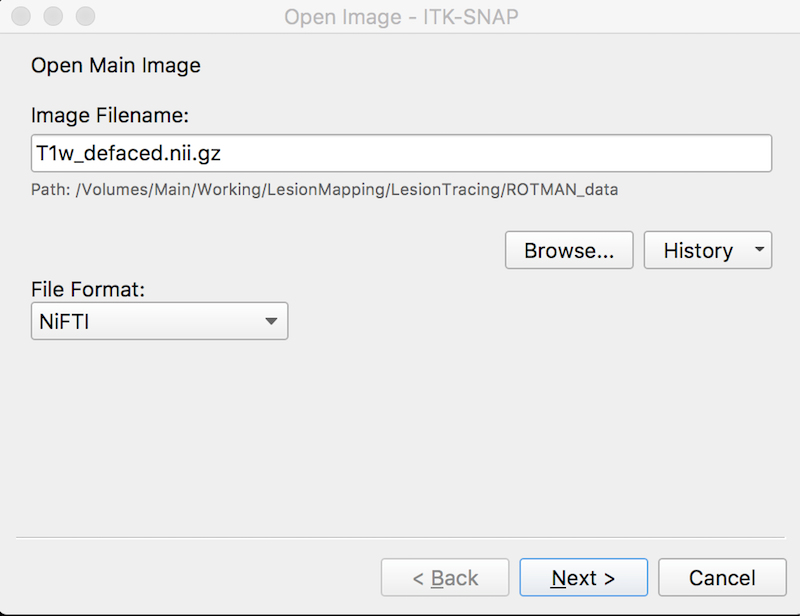
- Press Next. You see summary information and a warning about lack of precision. This is fine.
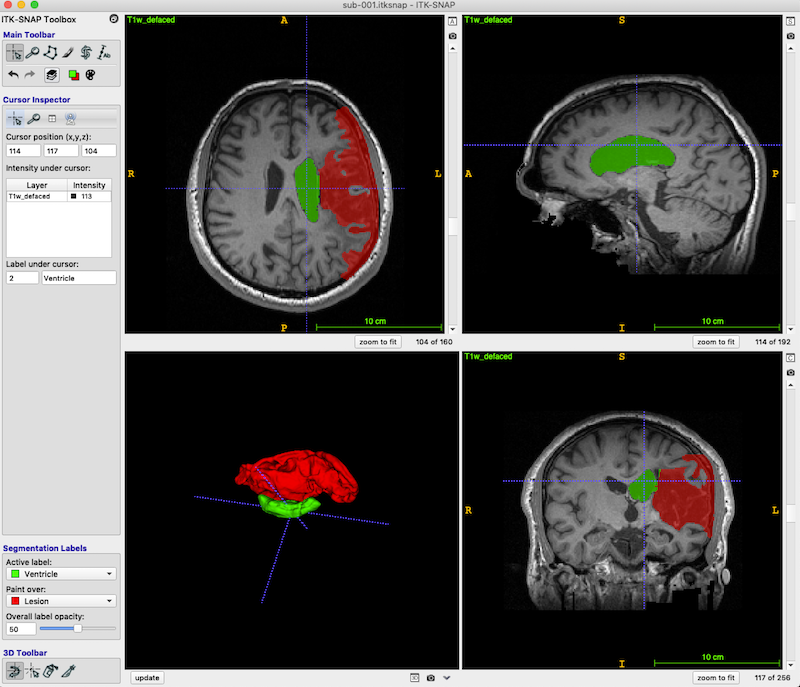
- Press Finish. ITK-SNAP’s main window should now display three orthogonal views of the MRI scan.
- If the images are too dark, adjust the contrast: Tools ➜ Image Contrast ➜ Auto Adjust Contrast (all layers).
- Press the zoom to fit button (red box, bottom right, in the figure below) so the head fills the available space.
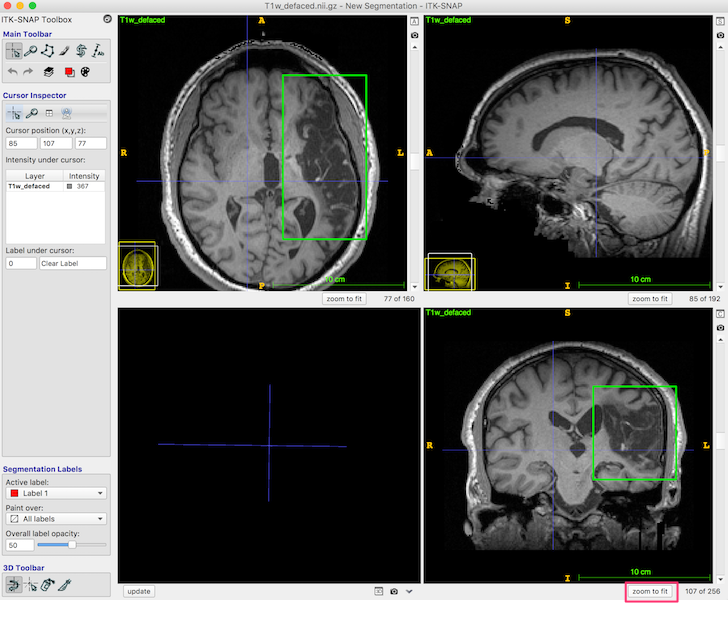
The dark area (enclosed in the green box in the above figure, axial and coronal views) is lesion. You should be able to distinguish both the CSF-filled region, and the area of damaged tissue around the rim. Some ventricle is unavoidably also included in the box.
If you have multiple images that would be useful for drawing the lesion, you can choose File Add Another Image. Snap will display all the images you have and yoke them together, even if they are different resolutions.
Understanding Labels¶
Segmentations (a.k.a. masks, labels) are simple images containing a single value. By default, most masks have the value 1 inside and 0 outside. However, if we want a mask that covers lesion and a different mask that covers ventricle, then we need masks with different values. For example, 1 could correspond to lesion and 2 could correspond to ventricle. Colors and names can be assigned to each label. ITK-SNAP has 6 default labels.
Label editor¶
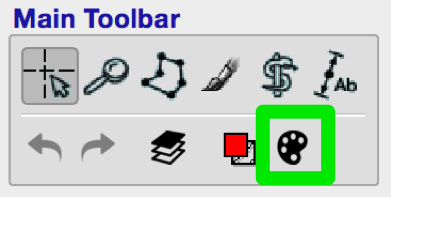
You can access the label editor by clicking on the palette icon in the Main Toolbar (green box, figure below).
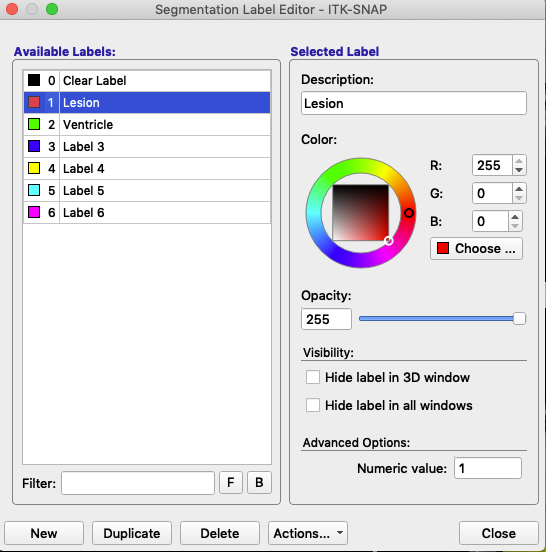
The label editor looks like this:
The label is defined by its Numeric value (lower right). Description and associated color (right, figure above) can be modified. In this figure, the label with the value 1 remains red, but has been renamed Lesion. The label with the value 2 remains green, but has been renamed Ventricle. At the bottom of the label editor, click Actions to see options to import or export a set of label descriptions.
Segmentation labels¶
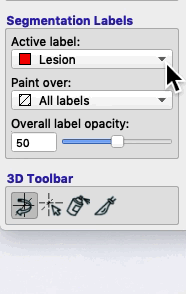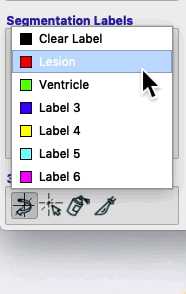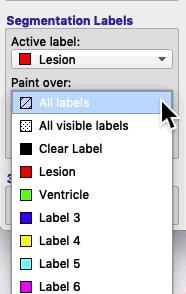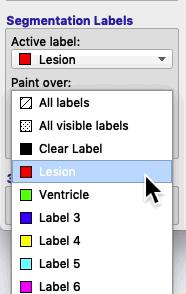
On the bottom left of the ITK-SNAP interface is the Segmentation Labels section. As shown in the animated gif below, you can select the active label (i.e. the label value that will be applied by any of the 2D editing tools or automatic segmentation) AND you can select Paint over.
- If you select Paint over ➜ All labels, then the active label will replace any value it finds.
- If you select Paint over ➜ All visible labels, then the active label will replace any other segmentation label (but not the clear label).
- If you select Paint over ➜ Clear label, then the active label will apply only to areas of the image where there are no visible labels.
- If you select Paint over ➜ Label 4, then the active label will replace any areas that currently contain label 4, but no other areas. For example, if you have a brain mask with the label 4, and you paint over label 4, then you will never accidentally paint over areas outside the brain, because areas outside the brain will have the clear label.
- You can reduce the opacity of a label (like the brain mask) without affecting its functionality (i.e. even if the brain mask is completely transparent, it still behaves the same way).
Warning
Be careful about the Active label and Paint over settings! The feature is powerful.
2D Segmentation Editing¶
- If you have created a workspace, for example by following the instructions here: ITK-SNAP Segmentation of Stroke-related Lesions page, load that workspace now (e.g. Workspace → Open Workspace).
- Any changes you make to the segmentation can be saved with the same name. Be cautious about changing the names.
Paintbrush¶
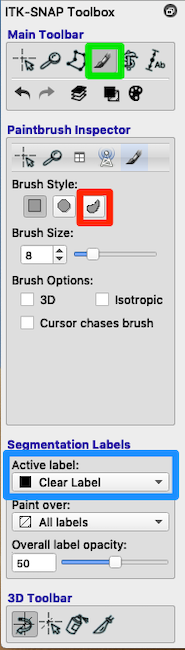
- Look at the toolbox. Choose the paintbrush (green box in above figure).
- Paint If you need to manually paint additional areas (e.g., parts of the lesion not filled by the algorithm), then set Active label = Lesion.
- Paint and Erase Mouse buttons: The left mouse button paints and the right mouse button erases.
- Erase with Clear Label If some of the lesion mask leaked into the ventricles you can use the paintbrush to erase it. Look at Segmentation Labels (blue box in figure). Painting with the Clear Label and Paint over: Lesion will erase lesion that has leaked into the ventricles.
Adaptive Brush¶
See the red square in the Paintbrush Inspector in the above figure. Try a relatively large brush size (e.g., 16 instead of 8), and try the 3D brush. Both of these choices should reduce the number of clicks you need to fill a region. The adaptive brush will paint everything that has an intensity similar to the intensity at the crosshairs AND is contiguous to the crosshairs. That is, if you have a large adaptive brush that straddles a tissue boundary, the fill will apply only to voxels contiguous to the crosshairs. The adaptive brush can be useful for filling in ventricles, which have a fairly consistent intensity, but very curvy borders. Click each time you select a new position for the crosshairs.
Note
Imagine that you have labeled the ventricle and you want to paint lesion right next to it without overwriting any of the ventricle. To preserve your ventricle, but paint lesion very close to it, choose Paint over = Clear Label instead of Paint over = All labels. The Clear Label is the default that covers all areas of the image where there is no visible label. Paint over = Clear Label will prevent the brush from overwriting another visible label.
Note
Overall label opacity (bottom of the figure) applies only to viewing the lesion segmentation label. It is nice to have this be translucent so you can see where to edit.
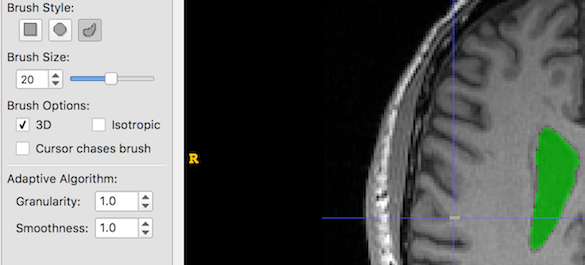
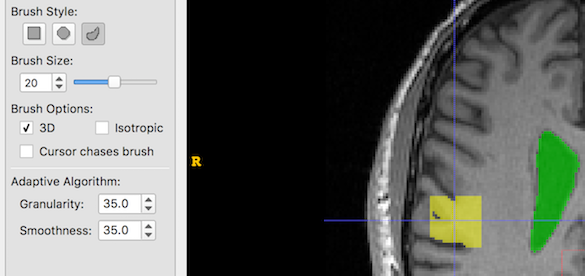
- The Adaptive Brush has two settings: Granularity and Smoothness. It is not immediately obvious what these mean, so below are some examples of creating a single brush stroke, size 20, at the same coordinates with different Granularity and Smoothness settings. Look at the yellow blob at the crosshair location:
First, we set Granularity and Smoothness to their minimums:
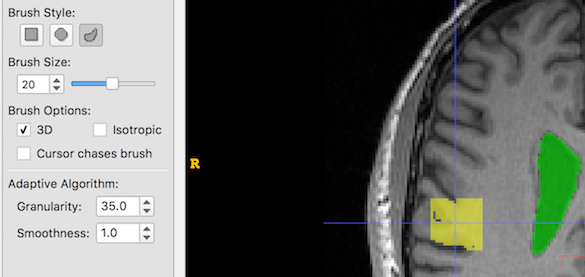
And now we increase Granularity only. Low granularity values lead to oversegmentation, while higher values lead to undersegmentation:
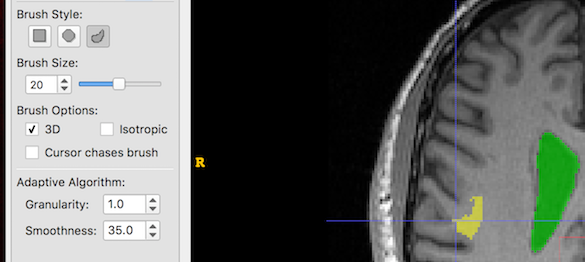
And now we increase Smoothness only. Larger smoothness values produce smoother segments:
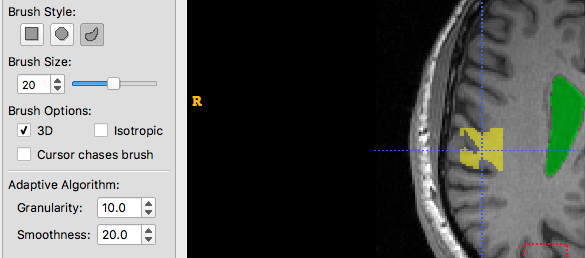
And now Granularity and Smoothness are both increased:
Finally, we try to find an appropriate balance:
Polygon¶
- Immediately to the left of the paintbrush on the Main Toolbar, is the polygon tool (green box in figure below).
- Select it and you’ll see the polygon inspector.
- Now you can draw around the structure of interest insuring that you are using the correct active label. To draw a polygon click the mouse to start, and then click again to define each line segment. You will need to close the figure.
Note
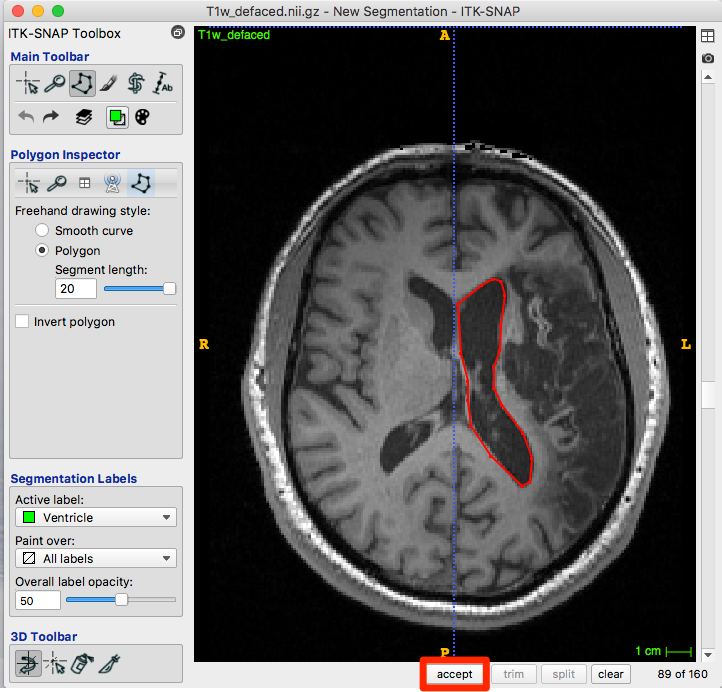
Although we use the ventricle label in the figure below, the polygon is drawn in the default red.
When you are happy with the polygon, click accept (red box, figure below).
After choosing accept, the roi is created with the appropriate color for your label:
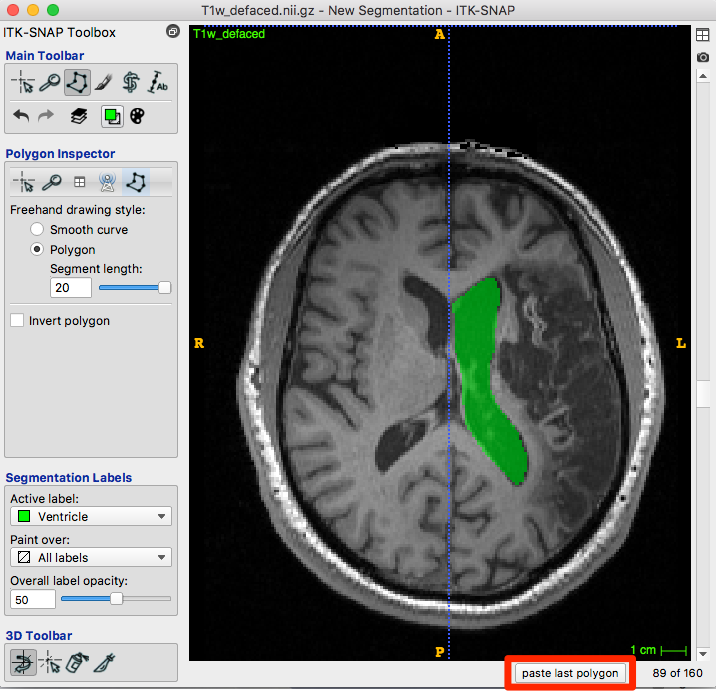
Below the image is a paste last polygon button (red box, figure below). Move to the next slice and click the button. A new polygon appears…you can drag vertices or edges, create new vertices or trim them until satisfied. Accept and repeat.
3D Segmentation Editing¶
3D editing tools only work once you have created a segmentation.
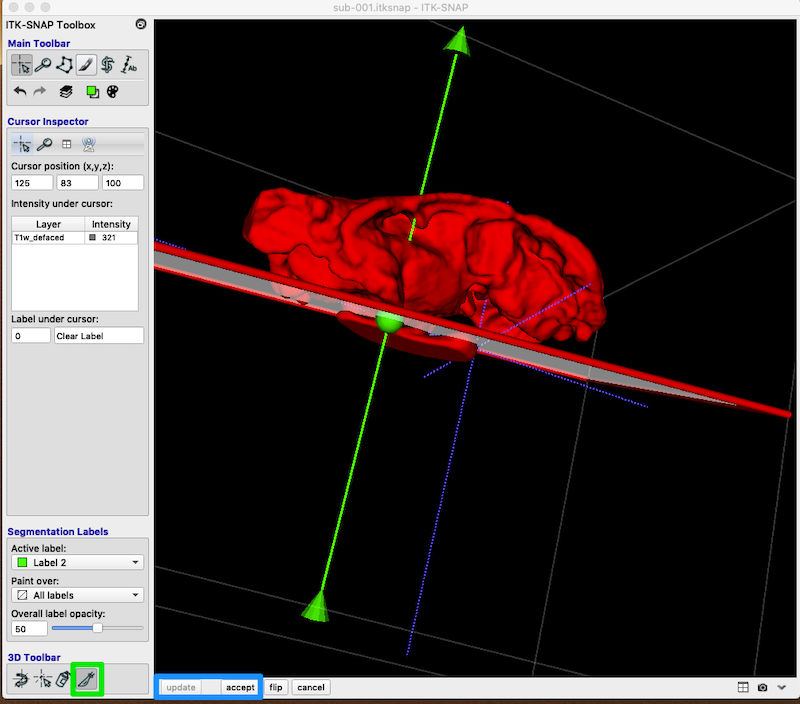
Scalpel¶
- You can create a plane through the 3D image and relabel values on the other side as something different.
- Set an active label other than the clear label.
- Identify a plane that roughly separates ventricle from lesion.
- Select the scalpel tool at the bottom of the toolbox (green box in figure below).
- You can manipulate the position of the cut plane with the arrow point, central ball or tail.
- When you are satisfied with the cut plane, click accept and update (blue box, bottom left).
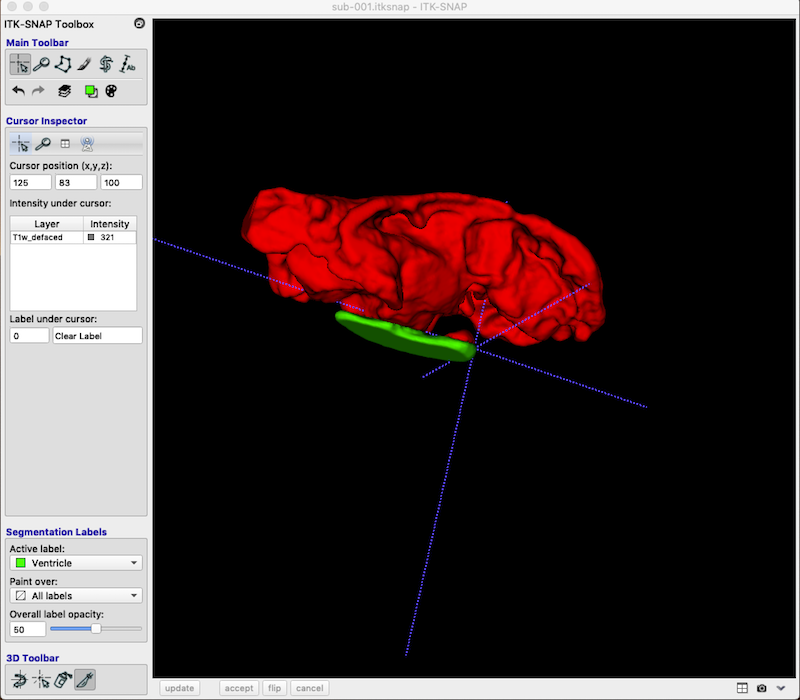
Your revision should look something like this (I have changed Label 2 to Ventricle):
Add a Second Segmentation¶
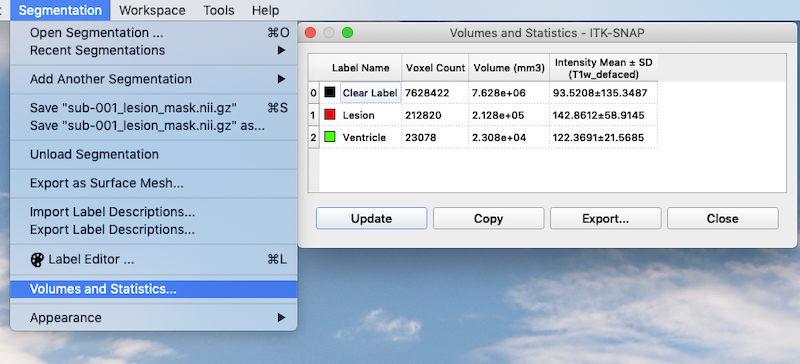
As you can see, you are not limited to a single segmentation type. Following the procedures above I have grown bubbles in the ventricle and added these to the final segmentation. Lesion is associated with the value 1. Ventricle is associated with the value 2.